Neural Networks from Scratch — part 4 Layers

Welcome! This is part 4 of Neural networks from scratch. In this guide, we’ll look into adding layers to our network. A layer is simply a bunch of neurons that get the same input. The output of each neuron is then sent together as the entire output of the layer.
Prerequisites
This part of the guide assumes that you already know how to multiply matrices together.
An adjustment to make our lives easier
A big problem with making neural networks is that we have so many different variables that are added and multiplied together. To make our lives easier, we can add a bunch of checks at the start of each function to ensure that our input is correct.
We’ll be doing this using the assert keyword. It works like this:
assert condition, error_emssageIf the condition is true, nothing happens. If the condition turns out to be false, the program will throw an Assertion Error with the error message we provide.
Before getting to these assertion checks, we’re changing a bit about the math. Remember how I said that X and W are vectors and we’re performing their dot product?

Well, the proper way of doing this is as follows:
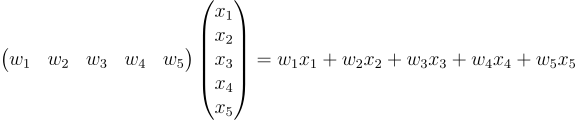
The first equation was the dot product. The second equation is actually a matrix product. This is preparing for what we’re going to do later. In the matrix product, the number of columns of the first matrix should be the same as the number of rows of the second matrix. This is extremely important. Reread that again if you’re not familiar with it. Our weights matrix is of shape 1x5. The input matrix is of shape 5x1. So those fives should match.
Here, the weight vector is called a row matrix and the input vector is called a column matrix.
At the start, we need to initialize our weights as a row matrix of the size 1xn, where n is the size of the input.
We do this as np.random.randn(1, input_size) . This function creates a random matrix with 1 row and input_size columns.
class Neuron:
def __init__(self, input_size):
assert isinstance(input_size, int), "Input size should be a number"
# Initializing random weight and bias
self.w = np.random.randn(1, input_size) * 0.01 # 1 x input_size
self.b = np.random.randn(1, 1) * 0.01 # 1 x 1This just checks if the input_size variable is an integer. We don’t need any more conditions for this function.
So, for 1xn shaped weight to be multiplied with the input, we need the input to have the shape of nx1. We’ll check that with the assert
def forward(self, x):
assert x.shape == (self.w.shape[1], 1)
self.input = x
self.z = np.dot(self.w, self.input) + self.b
assert self.z.shape == (1, 1), "There is a problem with forward process in neuron"
return self.zMoving to the backward function:
def backward(self, error, learning_rate):
# Getting the derivatives
assert error.shape == (1, ), f'Error sent to a neuron should be (1, ), but is {error.shape}'
dzdw = self.input.T
dzdb = 1
dzdprev = self.w.T
# These are the first and second terms of the gradient vector
dLdw = -error * dzdw # 1 x input_size
dLdb = -error * dzdb # 1 x 1
assert dLdw.shape == (1, self.input_size), 'dLdw is calculated incorrectly'
assert dLdb.shape == (1, ), f'dLdb is calculated incorrectly. It should be (1, ), but is {dLdb.shape}'
# Gradient clipping
dLdw = np.clip(dLdw, -1, 1)
dLdb = np.clip(dLdb, -1, 1)
# Updating our weight and bias
self.w += learning_rate * dLdw
self.b += learning_rate * dLdb
return dzdprevWe check that the error is just a single number. And finally, we also check if we’ve correctly calculated dLdw and dLdb based on their shapes.
Alright. Let’s make our layer class now
Layer Class
Our layer class should have a “neurons” variable. It will be the list of all the neurons in that layer.
class Layer:
def __init__(self, input_size, layer_size, activation):
self.neurons = [Neuron(input_size) for _ in range(layer_size)]
self.activation = activation
self.input_size = input_sizeWe take in the input and the layer size as paramters. For layer size, we make that many neurons. We also put in the input size to each neuron. We also take in the activation as a parameter. Finally, we just save the input size for later use.
Forward pass in the layer
def forward(self, input_vec):
self.z = np.array([n.forward(input_vec) for n in self.neurons]).reshape(-1, 1)
return self.activation.forward(self.z)This might appear slightly complicated so let’s break it down.
We take in the input vector as input_vec. Then we send it to each neuron. This will look like:
[n.forward(input_vec) for n in self.neurons]After sending input_vec to each neuron, we get the output and make a list out of that. But our problem is that we can’t send this array to the next layer. This is because the next layer expects the input to be a column vector. So, we first convert it into a NumPy array:
np.array([n.forward(input_vec) for n in self.neurons])Finally, we will reshape it to become a column vector. We do this in numpy as:
np.array([n.forward(input_vec) for n in self.neurons]).reshape(-1, 1)The array.reshape(-1, 1) works like this. The parameters we sent are -1 and 1. This tells Numpy the following: “Look, I want to convert this array into a matrix. The number of columns is 1, and you decide how many rows it should have”. The -1 is a wildcard. It would’ve worked just as well if we had put array.reshape(len(self.neurons), 1).
Finally, we run each element of the array through the activation function. Numpy handles that for us, so we just have to use: self.activation.forward(self.z)
Backwards pass of the layer
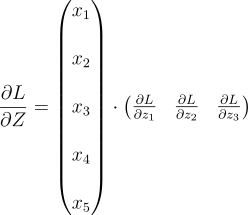
At each backwards pass of the layer, we expect dLdA . The derivative of the loss w.r.t the entire vector A. It will look like this:

Each element of the vector is the error for the respective neuron. So, all we gotta do is send the first element to the first neuron, the second element to the second neuron, and so on. But, each neuron expects dLdZ, not dLdA. So, we also need to multiply each element with the respective derivative of the activation function. If you remember this was the equation:
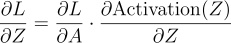
Since both Z and A are vectors, this equation is a vector equation. The full equation looks like this:
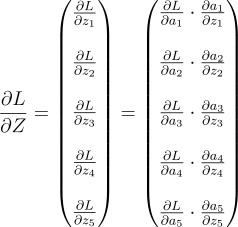
Note: This isn’t anything fancy. it’s the gradient descent for each neuron, but done parallely as a vector.
So, we have those vectors and we need to multiply them element-wise. We do this using np.multiply as follows:
def backward(self, dLdA, learning_rate):
# error size = layer_size x 1
assert dLdA.shape == (len(self.neurons), 1), f'Expected error to be of shape {(len(self.neurons), 1)}, but got {dLdA.shape}'
dLdZ = np.multiply(dLdA, self.activation.backward(self.z))Then we can send each element off to the respective neuron. But before that, we also need to get the gradient for the previous layer. In our neuron, this is named dzdprev.
Each neuron sends back this variable. It’s actually what the neuron thinks is the error of the previous layer. But since we have more than one neuron, we have multiple errors to send back. We simply add all of these errors and send dzdprev back.
Here’s the full code for the backwards pass:
def backward(self, dLdA, learning_rate):
# error size = layer_size x 1
assert dLdA.shape == (len(self.neurons), 1), f'Expected error to be of shape {(len(self.neurons), 1)}, but got {dLdA.shape}'
dLdZ = np.multiply(dLdA, self.activation.backward(self.z))
# Initialize dzdprev to be a bunch of zeros
dzdprev = np.zeros((self.input_size, 1))
for i, n in enumerate(self.neurons):
# Send the error to each neuron, get its output and add that to dzdprev
dzdprev = dzdprev + n.backward(dLdZ[i], learning_rate).reshape(-1, 1)
return dAnd that’s our layer class done! Here it is in all its glory:
class Layer:
def __init__(self, input_size, layer_size, activation):
self.neurons = [Neuron(input_size) for _ in range(layer_size)]
self.activation = activation
self.input_size = input_size
def forward(self, input_vec):
self.z = np.array([n.forward(input_vec) for n in self.neurons]).reshape(-1, 1)
return self.activation.forward(self.z)
def backward(self, dLdA, learning_rate):
# error size = layer_size x 1
assert dLdA.shape == (len(self.neurons), 1), f'Expected error to be of shape {(len(self.neurons), 1)}, but got {dLdA.shape}'
dLdZ = np.multiply(dLdA, self.activation.backward(self.z))
dzdprev = np.zeros((self.input_size, 1))
for i, n in enumerate(self.neurons):
dzdprev = dzdprev + n.backward(dLdZ[i], learning_rate).reshape(-1, 1)
return dzdprevNetwork class
Before this, we were storing all the neurons in our network class. Now we will just change that to store all layers. The logic is pretty much the same:
class Network:
def __init__(self, layers, loss_function):
self.layers = layers
self.loss_function = loss_function
def forward(self, a):
pred = a
for l in self.layers:
pred = l.forward(pred)
return pred
def backward(self, error, learning_rate):
for i in range(len(self.layers), 0, -1):
error = self.layers[i - 1].backward(error, learning_rate)
def train(self, training_data, training_labels, learning_rate, epochs):
for epoch in range(epochs):
total_error = 0
# Running the training
for i in range(len(training_data)):
pred = self.forward(training_data[i].reshape(-1, 1))
total_error += self.loss_function.forward(pred, training_labels[i].reshape(-1, 1))
error = self.loss_function.backward(pred, training_labels[i].reshape(-1, 1))
self.backward(error, learning_rate)
if epoch % (epochs/10) == 0:
print(f"epoch = {epoch}, error = {total_error/len(training_data)}")Testing our network
Let’s make this architecture:
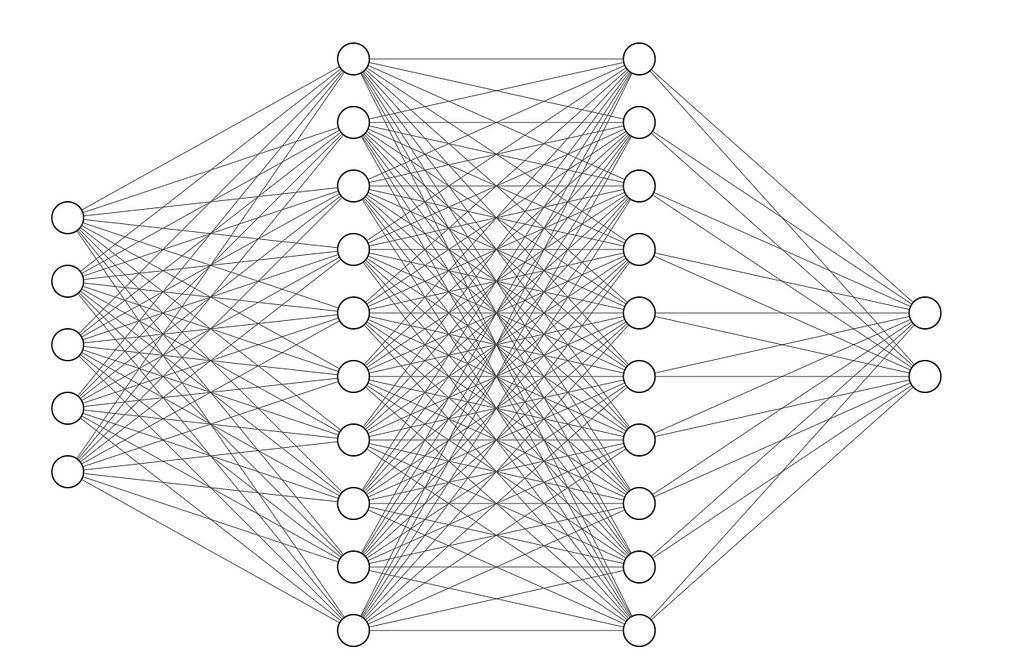
It takes in 5 elements. The first hidden layer has 10 neurons. the second hidden layer has 10 neurons. And finally, the last layer has 2 neurons. It looks like this:
n = Network(
[
Layer(window_size, 10, ReLU()),
Layer(10, 10, ReLU()),
Layer(10, forecast_size, Softmax()),
],
MSE(),
)
n.train(x, y, 0.0001, 1000)And running the code, we get:
epoch = 0, error = 0.04559152239976247
epoch = 100, error = 0.04559139774918803
epoch = 200, error = 0.04559135378732755
epoch = 300, error = 0.04559130825669434
epoch = 400, error = 0.04559126266737391
epoch = 500, error = 0.04559121896915248
epoch = 600, error = 0.04559117994778079
epoch = 700, error = 0.045591149103674615
epoch = 800, error = 0.04559113072754042
epoch = 900, error = 0.04559112984782191Although it is learning, it is learning VERY slowly. The errors are decreasing by about 0.00000014 or so. We can increase the learning rate to 0.001. But that is too big and our errors will start increasing. Also, this is taking a very long time to learn. Partly, it’s because of how many objects we have. We have 3 layers. The first layer has 10 neurons, the second has 10 and the third has 2. We have 22 objects floating around in our network. We have to store data in each object, call the functions update the data, and so on. If you notice, all that a layer class does is send data to each neuron. Maybe we can combine the neuron and layer data as one object.
Combining Layer and Neuron class
Suppose our layer has 3 neurons, each with 5 weights. Each neuron’s weights look like this:


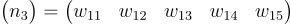
Instead of storing them as separate vectors, what if we store all this as a matrix?

Also, it’s kinda weird counting from 1 to 15 like this. Those numbers don’t make much sense. let’s name each element as Wij, where i represents the neuron and j represents the weight in that neuron.

We can also do the same for the biases and we get:
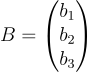
The forward pass will look like this now:
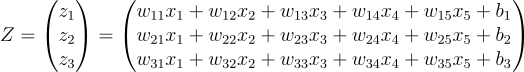
First of all, we can take out the last b values as its own vector

Then if you notice, the first term is just a matrix product of W and our input vector:

This can be written as an elegant equation:
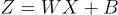
This is exactly like our old equation. But if you understood the previous explanation, you know that it is a lot more complex than our old equation.
This is the forward pass of 3 neurons with 10 weights each, all happening at the same time.
Let’s code this up before moving to the backwards pass:
class Layer:
def __init__(self, input_size, layer_size, activation):
self.shape = (input_size, layer_size)
self.W = np.random.randn(layer_size, input_size) * 0.001
self.B = np.random.randn(layer_size, 1) * 0.001
self.activation = activationOur initialized W and B variables are no longer vectors, but matrices.
def forward(self, X):
self.X = X
self.Z = np.dot(self.W, X) + self.B
self.A = self.activation.forward(self.Z)
return self.AThe forward pass is also the same. This is because Numpy handles both vectors and matrices in the backend, so we need to call the same function.
Backwards
This is where it can get slightly complicated. We will need to do some matrix algebra.
From dLdA to dLdZ
Each layer gets the dLdA error. This is a vector. Each element of the vector is the error for the respective neuron. We would first need to multiply it by dAdZ (This is the gradient of the activation function) to get dLdZ . Again, this is pretty much the same as what we had before. The backwards pass of the activation function returns another vector. We need to multiply both vectors element-wise. The code is:
dLdZ = np.multiply(dLdA, self.activation.backward(self.Z))At this point, if you’re wondering the difference between np.dot and np.multiply. np.multiply simply multiples the vectors element but np.dot also adds along the horizontal axis.
If I have two vectors A and B. np.multiply(A, B) will give me another vector where each element is the output of the multiplication of the respective elements of A and B. But np.dot(A, B) will give me a single number.
From dLdZ to dLdW
Here we have to use a trick. Remember that dLdZ is a column vector. It tells us the error of the output of each neuron.
Again suppose our layer only has 3 neurons with 5 weights each. Then, dLdZ will look like:
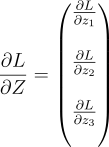
Take just the first element of it. We need to multiply that element with every element of the input vector (Remember that the gradient of Z w.r.t W is the input and the gradient of Z w.r.t the input is W. We will get:
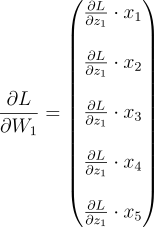
This is the gradient for the weights of just the first neuron. But, it’s not in the right shape. W1 is supposed to be a row vector. So we need to lay it out:

Similarly, we have to do that for the other two neurons as well:
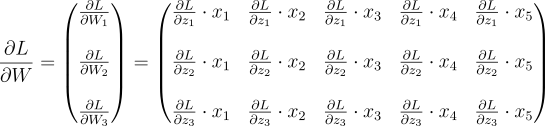
We have to get to that result using code.
Suppose I start by laying down dLdZ :
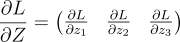
Then, I multiply our X vector with this:
What do you think the output will be? dLdz1 will get multiplied to each x, then dLdz2 will get multiplied to each x and finally, dLdz3 will get multiplied to each x. It will look like this:
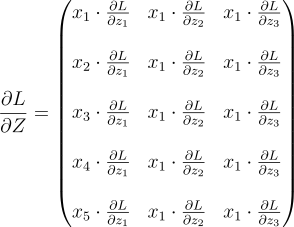
This is almost what we want. We want the transpose of this matrix.
We can do the above steps as follows:
- Laying down dLdZ : dLdZ.T
- Multiplying X with laid-down dLdZ : np.dot(self.X, dLdZ.T)
- Finally, taking the transpose: np.dot(self.X, dLdZ.T).T
From dLdZ to the previous layer’s error
We’re almost done. Remember that dLdB is just the same as dLdZ. We now have to worry about what to send back to the previous layer.
So, here’s what we have to do:
For each neuron, get the relevant value of dLdZ . Multiply that with every weight. You get a vector. That vector is what the neuron thinks is the error of the previous layer. You do this for each neuron. Finally, add all of the errors together to get the final error of the previous layer.
Let’s begin with our 3 neuron layer again. For the first neuron, we multiply dLdZ1 with each of its weights and we get a vector.
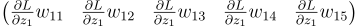
We do that for the other two neurons too:
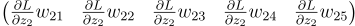

Then we add them together. But the problem is, they are row vectors. The previous layer is expecting a column vector. So before adding them, we need to transpose them:
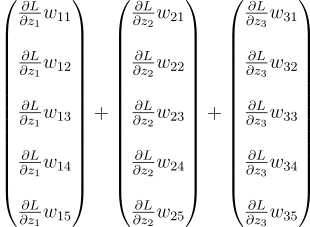
We have to get this. How do we do that? Well, let’s reverse-engineer it. Suppose we start from there. We start by adding all the vectors together:
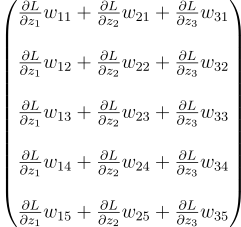
That looks like a matrix product. If I decompose it then it may be more apparent:
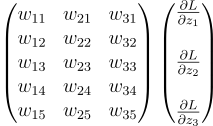
The first matrix is the transpose of the weights. And the secodn matrix is just dLdZ . So we can do this process with the line:
dLdA_prev = np.dot(self.W.T, dLdZ)Finally, we just have to multiply the gradients by the learning rate and subtract them from the weight matrix and the bias vector:
def backward(self, dLdA, learning_rate):
dLdZ = np.multiply(dLdA, self.activation.backward(self.Z))
dLdW = np.dot(self.X, dLdZ.T).T
dLdB = dLdZ
dLdA_prev = np.dot(self.W.T, dLdZ)
self.W -= learning_rate * dLdW
self.B -= learning_rate * dLdB
return dLdA_prevThe network class is much more simple now, since we’ve shifted most of the math to the layer class. I’ve modified some parts though. Instead of initializing the network class with the layers. We’ll initialize it empty. Then we can add the layers. The same is done for the loss function. Here is the class:
class Network:
def __init__(self):
self.layers = []
self.loss_function = None
def add_layers(self, layers):
self.layers.extend(layers)
def add_loss(self, loss_function):
self.loss_function = loss_function
def forward(self, X):
pred = X
for l in self.layers:
pred = l.forward(pred)
return pred
def backward(self, dLdA, learning_rate):
for l in reversed(self.layers):
dLdA = l.backward(dLdA, learning_rate)
def train(self, training_data, training_labels, learning_rate, epochs):
for epoch in range(epochs):
total_error = 0
for i in range(training_data.shape[1]):
pred = self.forward(training_data[i].reshape(-1, 1))
total_error += self.loss_function.forward(pred, training_labels[i].reshape(-1, 1))
err = self.loss_function.backward(pred, training_labels[i].reshape(-1, 1))
self.backward(err, learning_rate)
if epoch % (epochs // 5) == 0:
print(f'epoch = {epoch}, average error = {total_error / training_data.shape[1]}')Testing the network
n = Network()
n.add_layers([Layer(window_size, forecast_size, ReLU()), Layer(forecast_size, forecast_size, Identity())])
n.add_loss(MSE())
n.train(x, y, 0.0001, 10000)epoch = 0, average error = 0.3217097840344489
epoch = 2000, average error = 0.04422676149253911
epoch = 4000, average error = 0.039144083738589575
epoch = 6000, average error = 0.03905068719601368
epoch = 8000, average error = 0.03904892563272784It does seem to be learning. However, the errors are decreasing much slower too. Possible improvements are:
- Better gradient descent
- Better training (using batch processing)
Conclusion
In this guide, we expanded our neural network to incorporate layers, significantly enhancing its capability to learn complex patterns.
By understanding and implementing these foundational concepts, you now have the tools to build more sophisticated neural networks from scratch. Keep experimenting with different architectures and techniques to continue improving your models. Happy coding!